GLUCOSE DEHYDROGENASE (NAD(P)-dependent) from Microorganism
GLD-311
| Appearance: | White amorphous powder, lyophilized | |
|---|---|---|
| Activity: | GradeⅢ 250U/mg-solid or more | |
| Contaminants: | NADH oxidase ≤1.0×10⁻³% α-Glucosidase ≤1.0×10⁻³% Glucose-6-phosphate dehydrogenase ≤1.0×10⁻³% |
|
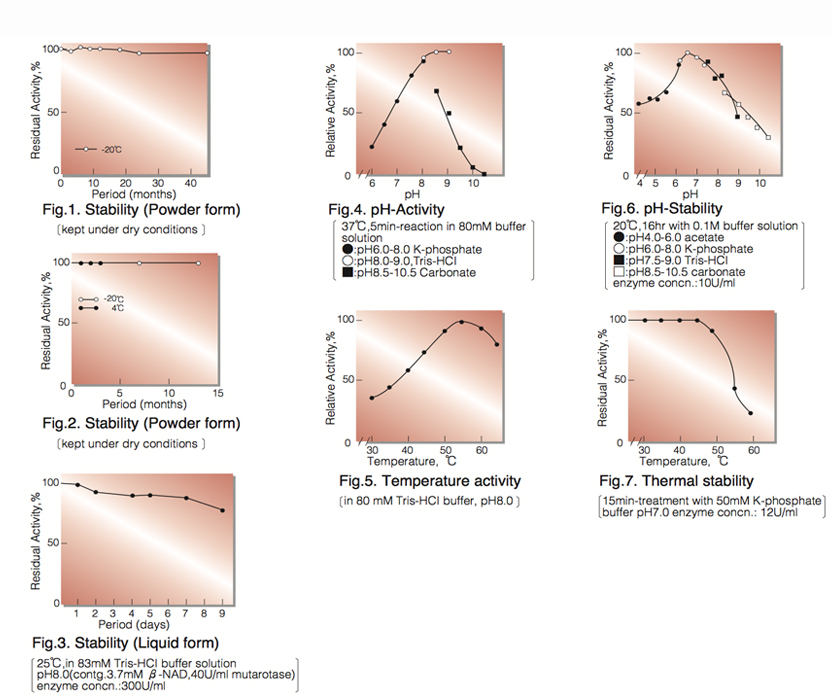
| Stability: | Stable at -20°C for at least One year (Fig.1) |
|---|---|
| Molecular weight: | approx. 101,000 (Gel filtration) |
| Isoelectric point: | 4.5 |
| Michaelis constants: | NAD⁺linked : 1.38×10⁻²M (D-Glucose) 3.09×10⁻⁴M (NAD⁺) NADP⁺linked : 1.25×10⁻²M (D-Glucose) 4.07×10⁻⁵M (NADP⁺) |
| Inhibitors: | Ag⁺, Hg²⁺, Monoiodoacetate |
| Optimum pH: | 9.0(Fig.4) |
| Optimum temperature: | 55℃(Fig.5) |
| pH Stability: | pH 6.0-7.5 (20℃, 16hr)(Fig.6) |
| Thermal stability: | 45℃ (15min-treatment with 50mM K-phosphate buffer, pH 7.0)(Fig.7) |
| Substrate specificty: | Specific for ß-D,-Glucose or 2-Deoxy-glucose (Table.1) (Either NAD⁺ or NADP⁺ serves as coenzyme.) |
APPLICATIONS
This enzyme is useful for enzymatic determination of D-Glucose.
ASSAY
Principle:
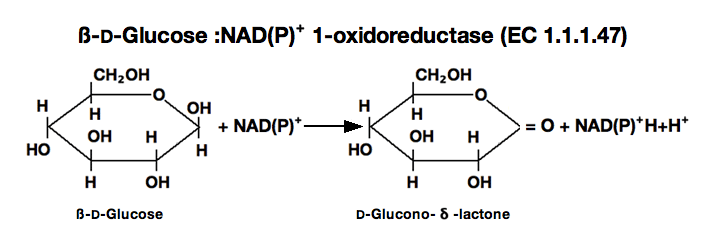
glucose dehydrogenase
ß-D-Glucose+NAD⁺ ►D-Glucono-δ-lactone+NADH+H⁺
The appearance of NADH is measured at 340nm by spectrophotometry.
Unit definition:
One unit causes the formation of one micromole of NADH per minute under the conditions described below.
Method:
| A. Tris-HCl buffer, pH 8.0: | 0.1M |
|---|---|
| B. D-Glucose solution: | 1.5M |
| C. ß-NAD⁺ solution: | 80mg/ml |
| D. Enzyme diluent: | 50mM K-phosphate buffer, pH 7.0 contg. 0.1% BSA |
Procedure
| Concentration in assay mixture | |
|---|---|
| Tris-HCl buffer | 85.25mM |
| D-Glucose | 147.54mM |
| NAD⁺ | 3.66mM |
1. Prepare the following reaction mixture in a cuvette (d=1.0cm) and equilibrate at 37℃ for about 5 minutes.
2.6 ml Tris-HCl buffer, pH 8.0 (A)
0.3ml Substrate solution (B)
0.1ml ß-NAD⁺ solution(C)
2. Add 0.05ml of the enzyme solution* and mix by gentle inversion
3. Record the increase in optical density at 340nm against water for 2 to 5 minutes in a spectrophotometer
thermostated at 37℃, and calculate theΔOD per minute from the initial linear portion of the curve (ΔOD test).
At the same time, measure the blank rate (ΔOD blank) by using the same method as the test except that the enzyme diluent (D) is added instead of the enzyme solution.
* Dissolve the enzyme preparation in ice-cold enzyme diluent (D), dilute to 0.8-1.2U/ml with the same buffer and store on ice.
Calculation
Activity can be calculated by using the following formula :

ΔOD/min (ΔOD test−ΔOD blank ) ×Vt × df
Volume activity (U/ml) = =ΔOD/min×9.807×df
6.22×1.0×Vs
Weight activity (U/mg)=(U/ml)×1/C
- Vt
- : Total volume (3.05ml)
- Vs
- : Sample volume (0.05ml)
- 6.22
- : Millimolar extinction coefficient of NADH under the assay conditions (㎠/micromole)
- 1.0
- : Light path length (cm)
- df
- : Dilution factor
- C
- : Enzyme concentration in dissolution (c mg/ml)
| Substrate (150mM) | Relative activity(%) | Substrate (150mM) | Relative activity(%) |
|---|---|---|---|
| D-Glucose | 100 | Galactose | 1.7 |
| L-Glucose | 0.0 | D-Lactose | 1.5 |
| D-Xylose | 16.2 | D-Sorbitole | 0.0 |
| 2-Deoxy-glucose | 127.0 | D-Mannitol | 0.0 |
| L-Sorbose | 0.0 | Sucrose | 0.0 |
| D-Mannose | 5.1 | Inositol | 0.0 |
| D-Fructose | 0.0 | Maltose | 1.4 |
| Chemical | Concn.(mM) | Residual activity(%) |
Chemical | Concn.(mM) | Residual activity(%) |
|---|---|---|---|---|---|
| None | − | 100 | KF | 2.0 | 98.7 |
| Metal salt | 2.0 | NaF | 10.0 | 100.6 | |
| AgNO₃ |
7.1 | NaN₃ | 20.0 | 101.6 |
|
| Ba(OAc)₂ | 98.2 | NEM | 2.0 | 97.6 | |
| CaCl₂ | 98.9 | MIA | 2.0 | 0.4 | |
| Cd(OAc)₂ | 96.6 | IAA |
2.0 | 92.2 | |
| CoCl₂ | 96.4 | EDTA | 5.0 | 107.2 | |
| CuSO₄ | 99.5 | (NH₄)₂SO₄ | 20.0 | 96.0 | |
| FeCl₃ | 98.1 | Borate | 20.0 | 101.4 | |
| FeSO₄ | 96.6 | o-Phenanthroline | 2.0 | 97.7 | |
| HgCl₂ | 5.9 | α,α′-Dipyridy | 1.0 | 100.3 | |
| MgCl₂ | 101.5 | Urea | 2.0 | 122.5 | |
| MnCl₂ | 100.9 | Guanidine | 2.0 | 99.2 | |
| NiCl₂ | 93.4 | Hydroxylamine | 2.0 | 107.2 | |
| Pb(OAc)₂ | 99.8 | ||||
| ZnSO₄ | 102.1 |
Ac, CH₃CO; PNEM, N-Ethylmaleimide; MIA, Monoiodoacetate; IAA, lodoacetamide; EDTA, Ethylenediaminetetraacetate.