Life Science

Code No. TRT-101 10,000U
*This product is not available in the US.
High Efficient Reverse Transcriptase
ReverTra Ace®
Description
ReverTra Ace® is a high efficient M-MLV (Moloney Murine Leukemia Virus) reverse transcriptase that has been genetically modified to remove RNase H activity and increase reaction efficiency. It is the preferred enzyme for applications requiring full-length cDNAs and high product yields from total RNA, mRNA, rRNA, etc.
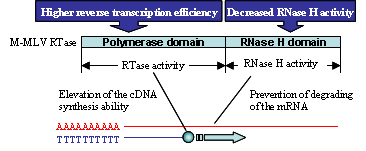
Features
- RNase minus M-MLV RTase with improved performance
- Enables the synthesis of longer cDNAs (≥ 14 kb) than the WT-enzyme
- Exhibits excellent reaction efficiency at high temperatures.
Applications
- cDNA synthesis
- cDNA library construction
- RT-PCR
- 5’ RACE
Source
E. coli strain carrying a cloned, modified M-MLV reverse transcriptase gene.
Unit definition
One unit is defined as the amount of enzyme required to incorporate 1 nmole of dTTP into an acid-insoluble material in 10 min at 42ºC.
Storage condition
50 mM Tris-HCl (pH 7.5), 100 mM NaCl, 0.1 mM EDTA, 10 mM DTT, 0.01% Nonidet® P-40, 50% Glycerol. Store at -20ºC
Components
ReverTra Ace® (100U/μl)
5 x Buffer
Typical PCR reaction setup
| Component | Amount | |
|---|---|---|
| RNA Total RNA Poly (A)+ RNA |
0.1-1 μg 50-500 ng |
<Reaction condition> 30ºC, 10 min** 42ºC, 20-60 min 99ºC, 5 min ** when using random primers |
| Primer Oligo (dT) Random Gene specific |
5 pmoles 25 pmoles 5 pmoles |
|
| 5 x Buffer | 4 μl | |
| 10 mM dNTPs* | 2 μl | |
| (RNase inhibitor ) | 20 U | |
| Nuclease-free water | X μl | |
| Total reaction volume | 20 μl | |
| * This reagent is not supplied with this product. | ||
| Component | Amount | |
|---|---|---|
| Poly (A)+ RNA | 50-500 ng |
<Reaction condition> 42ºC, 30 min 99ºC, 5 min |
| Oligo (dT) primer | 5 pmoles | |
| 5 x Buffer | 4 μl | |
| 10 mM dNTPs* | 2 μl | |
| (RNase inhibitor ) | 20 U | |
| Nuclease-free water | X μl | |
| Total reaction volume | 20 μl | |
| * This reagent is not supplied with this product. | ||
Application data
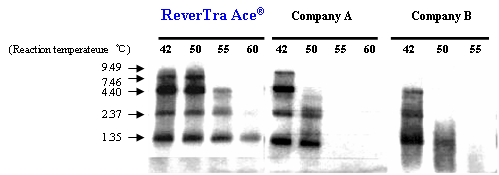
Example 1. Comparison of elongation capability of RNase H minus RTases at various temperatures
cDNAs were synthesized with oligo (dT)30 primers and 100 U enzyme/poly (A)+ RNA mixture (1.35-9.49 kb, 0.4 mg) as templates for 30 min at various temperatures. cDNAs were labeled with [32P-dCTP] during the reaction. The synthesized cDNAs were separated by 1% denatured agarose gel electrophoresis, and detected. The results suggested that ReverTra Ace® can elongate efficiently at 42-55ºC compared to other RNase H minus RTases from other companies.
Example 2. Comparison of cDNA synthesis efficiency by RT-PCR
G3PDH genes (500 bp) were amplified by PCR using cDNA templates that were synthesized with various RNase H minus RTases from G3PDH mRNA (102-105 copies/reaction). The RTase reaction was performed with specific reverse primers and 100 U enzyme at 42ºC for 20 min. The results indicated that ReverTra Ace® is suitable for RT-PCR amplifications that require sensitivity.

Example 3. Confirmation of the elongation capability of a long cDN
cDNA was synthesized by ReverTra Ace® using a specific primer for the 3’-end of dystrophin mRNA at 42ºC for 30 min. The 5’ region at a distance of 14 kb from the 3’ end of the dystrophin gene was amplified by PCR. The result indicated that ReverTra Ace® can elongate cDNA of ≥ 14 kb.

References
- K. Miyazaki, H. Miyamoto, D.K. Mercer, T. Hirase, J.C. Martin, Y. Kojima, H.J. Flint, Involvement of the multidomain regulatory protein XynR in positive control of xylanase gene expression in the ruminal anaerobe Prevotella bryantii B(1)4. J Bacteriol. 185: 2219-26 (2003)
- A. Nezu, A. Tanimura, T. Morita, K. Irie, T. Yajima, Y. Tojyo, Evidence that zymogen granules do not function as an intracellular Ca2+ store for the generation of the Ca2+ signal in rat parotid acinar cells. Biochem J. 363: 59-66 (2002)
- S. Nakamura, A. Ikegami, Y. Matsumura, T. Nakanishi, K. Nomura, Molecular cloning and expression of the mannose/glucose specific lectin from Castanea crenata cotyledons. J Biochem. 13: 241-6. (2002)
- S. Atsumi, Y. Ikawa, H. Shiraishi, T. Inoue, Design and development of a catalytic ribonucleoprotein. EMBO J. 2: 5453-60 (2001)
- Y. Yabuta, K. Yoshimura, T. Takeda, S. Shigeoka, Molecular characterization of tobacco mitochondrial L-galactono-gamma-lactone dehydrogenase and its expression in Escherichia coli. Plant Cell Physiol. 41: 666-75 (2000)
- S. Lee. Y. Takeda, H. Kawano, H. Hosoya, M. Nomoto, D. Fujimoto, N. Takahashi, K. Watanabe. Expression and regulation of a gene encoding neural recognition molecule NB-3 of the contactin/F3 subgroup in mouse brain. Gene, 245: 253-66 (2000)
To get a quote, contact us at info@toyobousa.com, or INQUIRY.